Introduction
On April 10th, 1912, the RMS Titanic departed on its maiden voyage from Southampton to New York City. After making stops in Cherbourg and Queenstown to pick up additional passengers, the ship headed towards New York. There were 2,224 passengers including 892 crew members aboard the ship. This was under the ship’s capacity of around 3,300. The ship was equipped with watertight compartments that were designed to fill up in the event of a breach to the ship’s hull. This technology was said to make the ship “unsinkable”.
The Titanic was scheduled to arrive at New York Pier 59 on the morning of April 17th. But at around 11:40 p.m. on the night of April 14th, the ship collided with an iceberg. The ship’s hull was not punctured by the collision, however it weakened the seams of the hull causing them to separate. Several watertight compartments filled with water and the ship eventually sank in the Atlantic Ocean about 400 miles off the coast of Newfoundland.
Distress signals were sent out, but no ships were near enough to reach the Titanic before she sank. At around 4 a.m., the the RMS Carpathia arrived on scene in response to the distress signals The ship had 20 lifeboats which had capacity for only 1,178 passengers. Approximately 1,500 people lost their lives in this tragic event. About 710 people survived and were taken to New York via the Carpathia.
So, who were the passengers that survived this tragedy? Fortunately, we have data which can give us insight into who survived and who perished. The goal is to use machine learning methods to identify patterns in the data to predict which passengers survived and which ones did not.
The Dataset
The Titanic dataset comes from kaggle. The training set contains 891 rows and 12 columns with one row per passenger. The test set includes 418 rows and 11 columns (the survived column is missing for the sake of the competition).
The columns of the test set are:
- PassengerId: Kaggle passenger id
- Survived: 1 = passenger survived, 0 = passenger did not survive
- Pclass: Ticket class
- Name: Full name of passenger including title
- Sex: Sex of passenger
- Age: Age of passenger
- SibSp: Number of siblings and spouses aboard the ship
- Parch: Number of parents and children aboard the ship
- Ticket: Ticket number
- Fare: Passenger fare
- Cabin: Cabin number
- Embarked: Port of embarkation
Goal of Analysis
sources:
- https://en.wikipedia.org/wiki/RMS_Titanic
- https://www.kaggle.com/c/titanic/data
Import Data
Load the standard Libraries
First, load the standard python libraries.
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
%matplotlib inline
Next, we import the training data from the train.csv file and save it as data. After the data is loaded into our jupyter notebook, we convert it into a pandas dataframe and inspect it.
# import train data and save it as data
data = pd.read_csv('train.csv')
# load data into a pandas dataframe and save as df
df = pd.DataFrame(data)
df.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
Data Exploration
Now that the data is loaded, we can begin exploration. The data has already been split into training and test sets. We will explore the training set and act as if the test set is not available to us at this time. This will allow us to get a better measure of the accuracy our model and avoid over fitting.
The describe function is used to view summaries of the numerical columns in the dataset. This also helps to identify columns with null values. Notice the count of Age compared to the other columns.
df.describe()
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| count | 891.000000 | 891.000000 | 891.000000 | 714.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 446.000000 | 0.383838 | 2.308642 | 29.699118 | 0.523008 | 0.381594 | 32.204208 |
| std | 257.353842 | 0.486592 | 0.836071 | 14.526497 | 1.102743 | 0.806057 | 49.693429 |
| min | 1.000000 | 0.000000 | 1.000000 | 0.420000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 223.500000 | 0.000000 | 2.000000 | 20.125000 | 0.000000 | 0.000000 | 7.910400 |
| 50% | 446.000000 | 0.000000 | 3.000000 | 28.000000 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 668.500000 | 1.000000 | 3.000000 | 38.000000 | 1.000000 | 0.000000 | 31.000000 |
| max | 891.000000 | 1.000000 | 3.000000 | 80.000000 | 8.000000 | 6.000000 | 512.329200 |
The corr method calculates the correlation between variables.
c = df.corr()
c
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| PassengerId | 1.000000 | -0.005007 | -0.035144 | 0.036847 | -0.057527 | -0.001652 | 0.012658 |
| Survived | -0.005007 | 1.000000 | -0.338481 | -0.077221 | -0.035322 | 0.081629 | 0.257307 |
| Pclass | -0.035144 | -0.338481 | 1.000000 | -0.369226 | 0.083081 | 0.018443 | -0.549500 |
| Age | 0.036847 | -0.077221 | -0.369226 | 1.000000 | -0.308247 | -0.189119 | 0.096067 |
| SibSp | -0.057527 | -0.035322 | 0.083081 | -0.308247 | 1.000000 | 0.414838 | 0.159651 |
| Parch | -0.001652 | 0.081629 | 0.018443 | -0.189119 | 0.414838 | 1.000000 | 0.216225 |
| Fare | 0.012658 | 0.257307 | -0.549500 | 0.096067 | 0.159651 | 0.216225 | 1.000000 |
# Get a count of all columns including text fields
df.count()
PassengerId 891
Survived 891
Pclass 891
Name 891
Sex 891
Age 714
SibSp 891
Parch 891
Ticket 891
Fare 891
Cabin 204
Embarked 889
dtype: int64
Explore Age
# Unique values for age
df.Age.unique()
array([22. , 38. , 26. , 35. , nan, 54. , 2. , 27. , 14. ,
4. , 58. , 20. , 39. , 55. , 31. , 34. , 15. , 28. ,
8. , 19. , 40. , 66. , 42. , 21. , 18. , 3. , 7. ,
49. , 29. , 65. , 28.5 , 5. , 11. , 45. , 17. , 32. ,
16. , 25. , 0.83, 30. , 33. , 23. , 24. , 46. , 59. ,
71. , 37. , 47. , 14.5 , 70.5 , 32.5 , 12. , 9. , 36.5 ,
51. , 55.5 , 40.5 , 44. , 1. , 61. , 56. , 50. , 36. ,
45.5 , 20.5 , 62. , 41. , 52. , 63. , 23.5 , 0.92, 43. ,
60. , 10. , 64. , 13. , 48. , 0.75, 53. , 57. , 80. ,
70. , 24.5 , 6. , 0.67, 30.5 , 0.42, 34.5 , 74. ])
# distribution of passengers by age
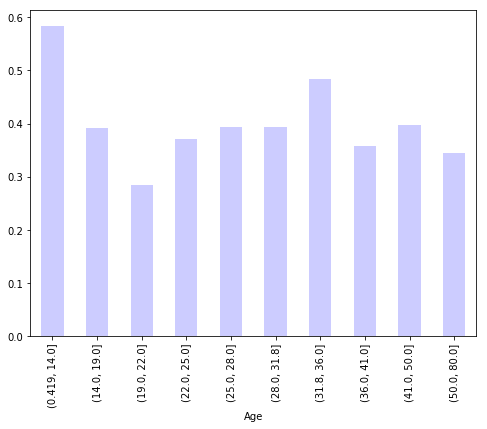
df.groupby(pd.qcut(df['Age'],10))['Survived'].mean().plot(figsize=(8,6), kind='bar', color='blue', alpha=.2)
Explore P Class
df['Pclass'].value_counts(dropna = False)
3 491
1 216
2 184
Name: Pclass, dtype: int64
# Survivors by Pclass
Explore Pclass
df.groupby('Pclass').describe()
| Age | Fare | ... | SibSp | Survived | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | count | mean | ... | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | |
| Pclass | |||||||||||||||||||||
| 1 | 186.0 | 38.233441 | 14.802856 | 0.92 | 27.0 | 37.0 | 49.0 | 80.0 | 216.0 | 84.154687 | ... | 1.0 | 3.0 | 216.0 | 0.629630 | 0.484026 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 |
| 2 | 173.0 | 29.877630 | 14.001077 | 0.67 | 23.0 | 29.0 | 36.0 | 70.0 | 184.0 | 20.662183 | ... | 1.0 | 3.0 | 184.0 | 0.472826 | 0.500623 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 3 | 355.0 | 25.140620 | 12.495398 | 0.42 | 18.0 | 24.0 | 32.0 | 74.0 | 491.0 | 13.675550 | ... | 1.0 | 8.0 | 491.0 | 0.242363 | 0.428949 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
3 rows × 48 columns
Survivors by Pclass
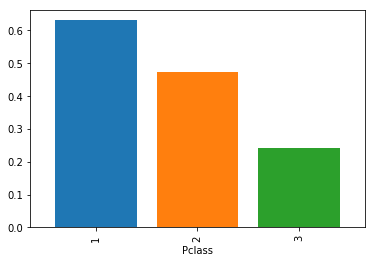
df.groupby('Pclass')['Survived'].mean()
Pclass
1 0.629630
2 0.472826
3 0.242363
Name: Survived, dtype: float64
df.groupby('Pclass')['Survived'].mean().plot(kind='bar',width=0.8)
Explore Embarked Location
df['Embarked'].value_counts(dropna = False)
S 644
C 168
Q 77
NaN 2
Name: Embarked, dtype: int64
pd.crosstab(df['Pclass'],df['Embarked'])
| Embarked | C | Q | S |
|---|---|---|---|
| Pclass | |||
| 1 | 85 | 2 | 127 |
| 2 | 17 | 3 | 164 |
| 3 | 66 | 72 | 353 |
Survivors by Embarked Location
df.groupby('Embarked')['Survived'].mean()
Embarked
C 0.553571
Q 0.389610
S 0.336957
Name: Survived, dtype: float64
df.groupby('Embarked')['Survived'].mean().plot(kind='bar',width=0.8)
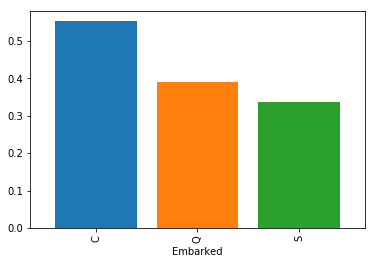
Feature Engineering
Age Binning
To make the age field more informative, we can group passengers into age bins.
df['age_bin'] = pd.qcut(df['Age'],10)
pd.crosstab(df['age_bin'],df['SibSp'])
| SibSp | 0 | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| age_bin | ||||||
| (0.419, 14.0] | 20 | 24 | 6 | 7 | 16 | 4 |
| (14.0, 19.0] | 60 | 20 | 3 | 1 | 2 | 1 |
| (19.0, 22.0] | 55 | 9 | 3 | 0 | 0 | 0 |
| (22.0, 25.0] | 48 | 15 | 5 | 2 | 0 | 0 |
| (25.0, 28.0] | 45 | 14 | 2 | 0 | 0 | 0 |
| (28.0, 31.8] | 47 | 18 | 0 | 1 | 0 | 0 |
| (31.8, 36.0] | 63 | 26 | 1 | 1 | 0 | 0 |
| (36.0, 41.0] | 34 | 17 | 2 | 0 | 0 | 0 |
| (41.0, 50.0] | 50 | 26 | 2 | 0 | 0 | 0 |
| (50.0, 80.0] | 49 | 14 | 1 | 0 | 0 | 0 |
pd.crosstab(df['Parch'],df['SibSp'])
| SibSp | 0 | 1 | 2 | 3 | 4 | 5 | 8 |
|---|---|---|---|---|---|---|---|
| Parch | |||||||
| 0 | 537 | 123 | 16 | 2 | 0 | 0 | 0 |
| 1 | 38 | 57 | 7 | 7 | 9 | 0 | 0 |
| 2 | 29 | 19 | 4 | 7 | 9 | 5 | 7 |
| 3 | 1 | 3 | 1 | 0 | 0 | 0 | 0 |
| 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 |
| 5 | 2 | 3 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
df['family_members']=df['Parch']+df['SibSp']
df.groupby('family_members')['Survived'].agg(['mean', 'size'])
| mean | size | |
|---|---|---|
| family_members | ||
| 0 | 0.303538 | 537 |
| 1 | 0.552795 | 161 |
| 2 | 0.578431 | 102 |
| 3 | 0.724138 | 29 |
| 4 | 0.200000 | 15 |
| 5 | 0.136364 | 22 |
| 6 | 0.333333 | 12 |
| 7 | 0.000000 | 6 |
| 10 | 0.000000 | 7 |
df.groupby('age_bin')['Survived'].mean()
age_bin
(0.419, 14.0] 0.584416
(14.0, 19.0] 0.390805
(19.0, 22.0] 0.283582
(22.0, 25.0] 0.371429
(25.0, 28.0] 0.393443
(28.0, 31.8] 0.393939
(31.8, 36.0] 0.483516
(36.0, 41.0] 0.358491
(41.0, 50.0] 0.397436
(50.0, 80.0] 0.343750
Name: Survived, dtype: float64
Replacing Null Values of Age
# Save median age of train set to use with transform of test set
med_age = df["Age"].median()
# Replace null values of age with median age
df["Age_Fill_Med"] = df["Age"].fillna(med_age)
Get Title From Name
df['Title'] = df['Name'].apply(lambda s: s.split(', ', 1)[1].split(' ',1)[0])
#s=df['Name'].str.split(', ', 1)
pd.crosstab(df['Title'],df['family_members'])
| family_members | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Title | |||||||||
| Capt. | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Col. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Don. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dr. | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Jonkheer. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lady. | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Major. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Master. | 0 | 3 | 15 | 4 | 2 | 9 | 3 | 3 | 1 |
| Miss. | 100 | 27 | 22 | 10 | 9 | 4 | 6 | 1 | 3 |
| Mlle. | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mme. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mr. | 397 | 68 | 35 | 6 | 1 | 5 | 1 | 1 | 3 |
| Mrs. | 20 | 59 | 27 | 9 | 3 | 4 | 2 | 1 | 0 |
| Ms. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rev. | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sir. | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| the | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
df.groupby('Title').describe()
| Age | Age_Fill_Med | ... | Survived | family_members | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | count | mean | ... | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | |
| Title | |||||||||||||||||||||
| Capt. | 1.0 | 70.000000 | NaN | 70.00 | 70.000 | 70.0 | 70.00 | 70.0 | 1.0 | 70.000000 | ... | 0.00 | 0.0 | 1.0 | 2.000000 | NaN | 2.0 | 2.0 | 2.0 | 2.00 | 2.0 |
| Col. | 2.0 | 58.000000 | 2.828427 | 56.00 | 57.000 | 58.0 | 59.00 | 60.0 | 2.0 | 58.000000 | ... | 0.75 | 1.0 | 2.0 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Don. | 1.0 | 40.000000 | NaN | 40.00 | 40.000 | 40.0 | 40.00 | 40.0 | 1.0 | 40.000000 | ... | 0.00 | 0.0 | 1.0 | 0.000000 | NaN | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Dr. | 6.0 | 42.000000 | 12.016655 | 23.00 | 35.000 | 46.5 | 49.75 | 54.0 | 7.0 | 40.000000 | ... | 1.00 | 1.0 | 7.0 | 0.571429 | 0.975900 | 0.0 | 0.0 | 0.0 | 1.00 | 2.0 |
| Jonkheer. | 1.0 | 38.000000 | NaN | 38.00 | 38.000 | 38.0 | 38.00 | 38.0 | 1.0 | 38.000000 | ... | 0.00 | 0.0 | 1.0 | 0.000000 | NaN | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Lady. | 1.0 | 48.000000 | NaN | 48.00 | 48.000 | 48.0 | 48.00 | 48.0 | 1.0 | 48.000000 | ... | 1.00 | 1.0 | 1.0 | 1.000000 | NaN | 1.0 | 1.0 | 1.0 | 1.00 | 1.0 |
| Major. | 2.0 | 48.500000 | 4.949747 | 45.00 | 46.750 | 48.5 | 50.25 | 52.0 | 2.0 | 48.500000 | ... | 0.75 | 1.0 | 2.0 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Master. | 36.0 | 4.574167 | 3.619872 | 0.42 | 1.000 | 3.5 | 8.00 | 12.0 | 40.0 | 6.916750 | ... | 1.00 | 1.0 | 40.0 | 3.675000 | 2.092569 | 1.0 | 2.0 | 3.0 | 5.00 | 10.0 |
| Miss. | 146.0 | 21.773973 | 12.990292 | 0.75 | 14.125 | 21.0 | 30.00 | 63.0 | 182.0 | 23.005495 | ... | 1.00 | 1.0 | 182.0 | 1.263736 | 1.999089 | 0.0 | 0.0 | 0.0 | 2.00 | 10.0 |
| Mlle. | 2.0 | 24.000000 | 0.000000 | 24.00 | 24.000 | 24.0 | 24.00 | 24.0 | 2.0 | 24.000000 | ... | 1.00 | 1.0 | 2.0 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Mme. | 1.0 | 24.000000 | NaN | 24.00 | 24.000 | 24.0 | 24.00 | 24.0 | 1.0 | 24.000000 | ... | 1.00 | 1.0 | 1.0 | 0.000000 | NaN | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Mr. | 398.0 | 32.368090 | 12.708793 | 11.00 | 23.000 | 30.0 | 39.00 | 80.0 | 517.0 | 31.362669 | ... | 0.00 | 1.0 | 517.0 | 0.441006 | 1.154239 | 0.0 | 0.0 | 0.0 | 0.00 | 10.0 |
| Mrs. | 108.0 | 35.898148 | 11.433628 | 14.00 | 27.750 | 35.0 | 44.00 | 63.0 | 125.0 | 34.824000 | ... | 1.00 | 1.0 | 125.0 | 1.528000 | 1.347495 | 0.0 | 1.0 | 1.0 | 2.00 | 7.0 |
| Ms. | 1.0 | 28.000000 | NaN | 28.00 | 28.000 | 28.0 | 28.00 | 28.0 | 1.0 | 28.000000 | ... | 1.00 | 1.0 | 1.0 | 0.000000 | NaN | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
| Rev. | 6.0 | 43.166667 | 13.136463 | 27.00 | 31.500 | 46.5 | 53.25 | 57.0 | 6.0 | 43.166667 | ... | 0.00 | 0.0 | 6.0 | 0.333333 | 0.516398 | 0.0 | 0.0 | 0.0 | 0.75 | 1.0 |
| Sir. | 1.0 | 49.000000 | NaN | 49.00 | 49.000 | 49.0 | 49.00 | 49.0 | 1.0 | 49.000000 | ... | 1.00 | 1.0 | 1.0 | 1.000000 | NaN | 1.0 | 1.0 | 1.0 | 1.00 | 1.0 |
| the | 1.0 | 33.000000 | NaN | 33.00 | 33.000 | 33.0 | 33.00 | 33.0 | 1.0 | 33.000000 | ... | 1.00 | 1.0 | 1.0 | 0.000000 | NaN | 0.0 | 0.0 | 0.0 | 0.00 | 0.0 |
17 rows × 72 columns
df['Title'].value_counts()>10
Mr. True
Miss. True
Mrs. True
Master. True
Dr. False
Rev. False
Mlle. False
Col. False
Major. False
Jonkheer. False
Mme. False
Sir. False
Don. False
Ms. False
Capt. False
Lady. False
the False
Name: Title, dtype: bool
frequencies = df['Title'].value_counts()
condition = frequencies<10 # you can define it however you want
mask_obs = frequencies[condition].index
mask_dict = dict.fromkeys(mask_obs, 'Other')
df['New_Title'] = df['Title'].replace(mask_dict)
title_dict=df.groupby('New_Title')['Age'].median().to_dict()
idx=df['Age'].isnull()
# df.loc[idx,"Age"]=
df['Age2']=df['Age'].copy()
df.loc[idx,'Age2']=df.loc[idx,"New_Title"].map(title_dict)
df.loc[idx,['New_Title','Age','Age2']].head()
| New_Title | Age | Age2 | |
|---|---|---|---|
| 5 | Mr. | NaN | 30.0 |
| 17 | Mr. | NaN | 30.0 |
| 19 | Mrs. | NaN | 35.0 |
| 26 | Mr. | NaN | 30.0 |
| 28 | Miss. | NaN | 21.0 |
Replace Null Values of Fare
fare_median = df["Fare"].median()
fare_median
14.4542
Creating Dummy Variables for Embarked and Sex
from sklearn import preprocessing
df = pd.get_dummies(df,columns=['Embarked','Sex'],drop_first=True)
le = preprocessing.LabelEncoder()
le.fit(["C", "Q", "S"])
list(le.classes_)
['C', 'Q', 'S']
Fit the Model
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
y = df['Survived']
X = df[['Age_Fill_Med','Pclass','Sex_male','Fare']]
Random Forest Model and Cross Validation
# Original Random Forest with Eyal
rf = RandomForestClassifier(n_estimators=20,random_state=0)
# # Random Forest with Null Ages replaced with Median of Age
s1 = cross_val_score(rf,X,y,cv=10)
print('X: '+'{:.2f}% +- {:.2f}%'.format(100*np.mean(s1),100*np.std(s1)))
X: 80.93% +- 4.03%
Randomized Grid Search
Grid Search will test various hyperparameters and choose the best values for each. With a Random Forest classifier we have 3 main hyperparameters, max depth, max features, and min samples split.
Max depth is the depth of each tree in the forest. The depth is the number of splits a tree has. For example, setting max_depth = 3 will limit the number of splits per tree to 3.
Max features is the number of features in each tree. Limiting the number of features will prevent overfitting.
Min samples split is the number of records required to split on a given node. For example, the model with not split again if the number of records is less than this parameter.
from sklearn.model_selection import RandomizedSearchCV
from time import time
from scipy.stats import randint as sp_randint
def report(results, n_top=3):
for i in range(1, n_top + 1):
candidates = np.flatnonzero(results['rank_test_score'] == i)
for candidate in candidates:
print("Model with rank: {0}".format(i))
print("Mean validation score: {0:.3f} (std: {1:.3f})".format(
results['mean_test_score'][candidate],
results['std_test_score'][candidate]))
print("Parameters: {0}".format(results['params'][candidate]))
print("")
# specify parameters and distributions to sample from
param_dist = {"max_depth": [3, None],
"max_features": sp_randint(1,5),
"min_samples_split": sp_randint(2, 11),
"bootstrap": [True, False],
"criterion": ["gini", "entropy"],
"n_estimators": sp_randint(10,100)}
# run randomized search
n_iter_search = 30
random_search = RandomizedSearchCV(rf, param_distributions=param_dist,
n_iter=n_iter_search, cv=5)
start = time()
random_search.fit(X, y)
print("RandomizedSearchCV took %.2f seconds for %d candidates"
" parameter settings." % ((time() - start), n_iter_search))
report(random_search.cv_results_)
RandomizedSearchCV took 9.50 seconds for 30 candidates parameter settings.
Model with rank: 1
Mean validation score: 0.835 (std: 0.021)
Parameters: {'bootstrap': True, 'criterion': 'entropy', 'max_depth': None, 'max_features': 1, 'min_samples_split': 8, 'n_estimators': 74}
Model with rank: 2
Mean validation score: 0.831 (std: 0.029)
Parameters: {'bootstrap': True, 'criterion': 'gini', 'max_depth': None, 'max_features': 3, 'min_samples_split': 8, 'n_estimators': 76}
Model with rank: 3
Mean validation score: 0.824 (std: 0.020)
Parameters: {'bootstrap': True, 'criterion': 'entropy', 'max_depth': None, 'max_features': 3, 'min_samples_split': 7, 'n_estimators': 60}
rf = random_search.best_estimator_.fit(X,y)
rf.predict(X)
array([0, 1, 1, 1, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0,
1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1,
1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 1,
1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 1, 0, 0,
1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0,
0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0,
0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0,
0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1, 1, 1, 0, 0,
1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0,
0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1,
0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0,
1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1,
0, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 1,
1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 0,
0, 1, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0,
0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1,
0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 0, 0,
1, 0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0,
0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1,
1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 1, 0,
1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 1, 0,
0, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1,
1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1,
1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1,
0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 1,
0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0,
0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1,
0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0,
0, 0, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0,
1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1,
0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0,
0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0,
0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 1, 0, 0, 0, 1,
0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1, 1, 1,
1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1,
1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0], dtype=int64)
pd.DataFrame(rf.feature_importances_, index = X.columns
, columns = ['Feature Importance']).sort_values(by = 'Feature Importance', ascending = False)
| Feature Importance | |
|---|---|
| Fare | 0.330700 |
| Age_Fill_Med | 0.280745 |
| Sex_male | 0.280466 |
| Pclass | 0.108088 |
# run rf.predict() on the same columns as train data but using the test data once transformations are complete
Test model using the test dataset
Import Test data
Import test.csv and save it to dataframe test_df
test = pd.read_csv('test.csv')
test_df = pd.DataFrame(test)
test_df.head()
| PassengerId | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 892 | 3 | Kelly, Mr. James | male | 34.5 | 0 | 0 | 330911 | 7.8292 | NaN | Q |
| 1 | 893 | 3 | Wilkes, Mrs. James (Ellen Needs) | female | 47.0 | 1 | 0 | 363272 | 7.0000 | NaN | S |
| 2 | 894 | 2 | Myles, Mr. Thomas Francis | male | 62.0 | 0 | 0 | 240276 | 9.6875 | NaN | Q |
| 3 | 895 | 3 | Wirz, Mr. Albert | male | 27.0 | 0 | 0 | 315154 | 8.6625 | NaN | S |
| 4 | 896 | 3 | Hirvonen, Mrs. Alexander (Helga E Lindqvist) | female | 22.0 | 1 | 1 | 3101298 | 12.2875 | NaN | S |
# Fill NAs with median of training set
test_df["Age_Fill_Med"] = test_df["Age"].fillna(med_age)
test_df["Fare"] = test_df["Fare"].fillna(fare_median)
# Convert Embarked and Sex columns to dummy variables
test_df = pd.get_dummies(test_df,columns=['Embarked','Sex'])
# Remove unnecessary columns
test_df.drop(['Sex_female', 'Embarked_C' ], axis = 1, inplace = True)
# Explore columns for model
df[['Age_Fill_Med','Pclass','Sex_male','Fare']].describe()
| Age_Fill_Med | Pclass | Sex_male | Fare | |
|---|---|---|---|---|
| count | 891.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 29.361582 | 2.308642 | 0.647587 | 32.204208 |
| std | 13.019697 | 0.836071 | 0.477990 | 49.693429 |
| min | 0.420000 | 1.000000 | 0.000000 | 0.000000 |
| 25% | 22.000000 | 2.000000 | 0.000000 | 7.910400 |
| 50% | 28.000000 | 3.000000 | 1.000000 | 14.454200 |
| 75% | 35.000000 | 3.000000 | 1.000000 | 31.000000 |
| max | 80.000000 | 3.000000 | 1.000000 | 512.329200 |
# Add survival prediction to the test_df dataframe
test_df['Survived'] = rf.predict(test_df[['Age_Fill_Med','Pclass','Sex_male','Fare']])
print('Train survival mean: ' + str(round(df['Survived'].mean(),4)))
print('Test survival rate: ' +str(round(test_df['Survived'].mean(),4)))
Train survival mean: 0.3838
Test survival rate: 0.3278
Create and save submission file
submission = test_df[['PassengerId', 'Survived']]
submission.head()
| PassengerId | Survived | |
|---|---|---|
| 0 | 892 | 0 |
| 1 | 893 | 0 |
| 2 | 894 | 0 |
| 3 | 895 | 0 |
| 4 | 896 | 1 |
# Save submission to csv
# submission.to_csv('AR_Titanic_Submission.csv')
Conclusion
In conclusion, we see that the most important features to surviving the Titanic are age, gender, and fare.
